Simulating Correlated Likert Scale Data
Simulating Correlated Likert-Scale Data In R: 3 Simple Steps
Let us simulate some data!
Imagine you have sent out a questionnaire measuring a Construct using six items on a 7-point likert-scale.
You’re impatiently waiting for the responses and you want to start creating your data analysis code so you only have to insert the gathered data once it arrives. To make this process smooth, you want to have some realistic simulation of your actual data.
In the following lines of code, I will show you how to simulate the ideal scenario of intercorrelated Likert-Scale answers that measure one underlying factor (construct).
Loading the libraries
Let’s start by loading the libraries.
library(polycor) # for polychoric correlations
library(psych) # for scree plots
library(tidyverse)
library(faux) # for the function rnorm_pre
library(truncnorm) # for the function rtruncnorm
Creating simulation data
Step 1: Create a single item
The package truncnorm provides a function to sample data from a normal
distribution, but with boundaries (“truncated”)! This is just what you
want, as your data will be on a 7-point scale. a and b will be the
truncation limits. As I want to center my values around a mean of 0, I
will use -3 and 3. To get plausible likert-scale integers, I just use
the round function. As it’s sampled data, it doesn’t really matter if
the mean is not exactly zero.
##### SIMULATE DATA ######
# simulate NPI_1
NPI_1 <- round(rtruncnorm(n = 500,
mean = 0,
sd = 1,
a = -3,
b = 3))
head(NPI_1)
## [1] 0 -1 1 2 -1 0
mean(NPI_1)
## [1] -0.028
Step 2: Get its correlated cousins
Now we can use the rnorm_pre function from the faux package to
simulate data that is correlated to our first item. For this, we set the
same parameters (mu is the mean) as our original item and set the
correlations (r) to medium to strong correlations. Again, we use the
round function to make it a likert-scale data. In the end, we can
combine these data in a data.frame.
# get correlated NPI_2:
NPI_2 <- round(rnorm_pre(NPI_1, mu = 0, sd = 1, r = 0.7))
NPI_3 <- round(rnorm_pre(NPI_1, mu = 0, sd = 1, r = 0.6))
NPI_4 <- round(rnorm_pre(NPI_1, mu = 0, sd = 1, r = 0.7))
NPI_5 <- round(rnorm_pre(NPI_1, mu = 0, sd = 1, r = 0.8))
NPI_6 <- round(rnorm_pre(NPI_1, mu = 0, sd = 1, r = 0.75))
NPI <- tibble(NPI_1, NPI_2, NPI_3, NPI_4, NPI_5, NPI_6)
head(NPI)
## # A tibble: 6 x 6
## NPI_1 NPI_2 NPI_3 NPI_4 NPI_5 NPI_6
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0 -1 1 0 0 1
## 2 -1 0 -2 -1 -1 0
## 3 1 0 0 1 1 1
## 4 2 1 1 1 2 3
## 5 -1 -2 -1 1 0 -1
## 6 0 1 0 0 1 1
Step 3: Check the assumptions
Great! Now that we have our simulated data from six items, we want to
check whether they all load on one underlying latent factor. For this,
we can use polychoric correlations, but Pearson correlations might be
applicable too, see the book Modern Psychometrics in
R.
The hetcor function from the polycor package calculates
“A detailed study of this phenomenon within the context of structural equation models can be found in Rhemtulla et al. (2012). The authors suggest that in the area of 6, 7, or more categories, differences between Pearson and polychoric become negligible.”
See also here to learn how polychoric correlations work.
cormat <- hetcor(NPI) # calculate correlation matrix
eigen_values <- eigen(cormat)
scree(cormat$correlations, factors = FALSE) # nice
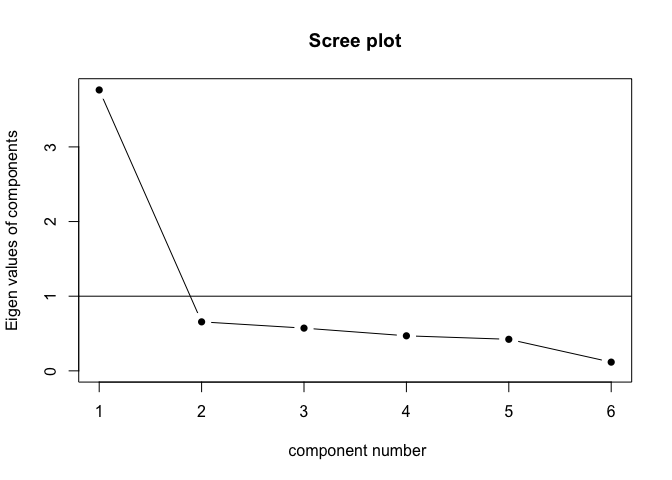
Cool! We actually seem to have one underlying component, as the “elbow” of the scree plot is behind the first component - see here for an explanation of scree plots.
Now let’s check the internal consistency using a custom-made cronbach’s alpha function:
# check internal consistency (cronbach's alpha):
cronbach.alpha <- function(data){
k <- ncol(data)
vcmat <- cov(data, use = "pairwise")
sigma2_Xi <- sum(diag(vcmat))
sigma2_X <- sum(vcmat)
alpha <- k/(k-1)*(1-sigma2_Xi/sigma2_X)
return(alpha)
}
cronbach.alpha(NPI)
## [1] 0.8777856
We seem to have a good internal consistency of the used scale! Horray!
You can now start using the data for your investigations, as if you’re using real data!
